Biologics that inhibit the pro-inflammatory cytokine IL-23 have long been regarded as important therapies in the treatment of psoriasis. However, much is still unknown about the molecular mechanisms that drive clinical efficacy. In this study, researchers sought to uncover the key driving factors by analyzing several IL-23 inhibitors in the treatment of psoriasis. Their findings, which are outlined in this post, shed light on the intricate interconnection between the molecular structure of IL-23 inhibitors and their clinical efficacy in treating psoriasis. This furthers our understanding of why some treatments may be more effective than others, allowing for optimal patient outcomes as individualized therapeutic approaches emerge as a possibility.
Psoriasis, a chronic autoimmune skin condition, presents both patients and providers with significant challenges in treatment and management. As it affects up to 40 million people worldwide, it has never been more critical to advance research and find effective solutions to help patients better manage their symptoms and work towards a cure.
Interleukin-23 (IL-23), known to be a dimeric cytokine consisting of two subunits, p19 and p40, plays a pivotal role in driving the inflammatory processes behind psoriasis. Current IL-23 inhibitor biologic treatments such as risankizumab, tildrakizumab, and guselkumab bind specifically to the p19 subunit, making them different mechanistically from other IL-23 inhibitor treatments like ustekinumab, which binds to the p40 subunit.
Prior research of the binding sites/epitopes of the anti-IL-23 biologics identified possible links between epitope structure and clinical efficacy, yet results were limited to ustekinumab as structural data were lacking for the p19-targeting biologics. As a result, the structural mechanisms of action of the p19-specific IL-23 inhibitors remained unclear.
This new groundbreaking study by Stefano G. Daniele, Sherif A. Eldirany, Giovanni Damiani, Minh Ho, and Christopher G. Bunick, “Structural Basis for p19 Targeting by Anti–IL-23 Biologics: Correlations with Short- and Long-Term Efficacy in Psoriasis,” delved into the epitope data for risankizumab, tildrakizumab, guselkumab, as well as ustekinumab, with the aim of characterizing the molecular and structural properties of IL-23 inhibitors and their impact on patient care.
"After caring for psoriasis patients for a long time using anti-IL-23 biologics, I wanted to know more about the unique chemistry and molecular structure governing the functional differences between them, especially the p19-specific biologics. That is how this project was born. I am a strong believer that structure dictates function, and by investigating this relationship I believe we can discover new knowledge that helps clinicians understand better the medicines they prescribe - both in mechanism and clinical efficacy and safety - and enables improved patient-tailored care. I hope this study is a springboard for more molecular and structural translational dermatology."
Dr. Christopher Bunick
Key Findings
The study revealed that while risankizumab, guselkumab, and tildrakizumab all bind exclusively to the p19 subunit, they do not necessarily do so in the exact same location on the subunit. This is reflected in the varying epitope surface areas (SA) which are occupied by each drug. Risankizumab was found to have the largest epitope SA, followed by guselkumab, ustekinumab, and lastly, tildrakizumab with the smallest epitope SA.
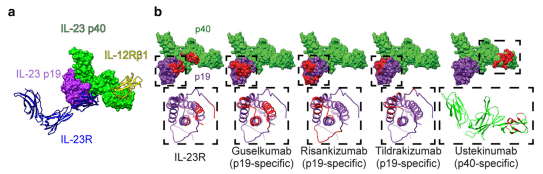
Variations were also noted for the degree of overlap between the drug’s epitope and the binding site for the IL-23 receptor (IL-23R). Guselkumab showed the highest overlap at 58%, risankizumab demonstrated an 8% overlap, and tildrakizumab a 0% overlap. Given the high clinical efficacy of risankizumab, this data suggests that the drug does not necessarily need to directly compete with IL-23R for the IL-23R binding site, but rather steric effects from the biologic are sufficient to disrupt IL-23R binding to IL-23.
The study further examined the chemical properties of the drug epitopes, comparing them to the binding site of the IL-23R, and found that the drug epitopes exhibited a more hydrophobic character compared to IL-23R binding site.
In examining the surface charge characteristics, the researchers found that risankizumab and ustekinumab had strongly acidic epitope surfaces compared to guselkumab and tildrakizumab which demonstrated a net neutral charge.
These variations in hydrophobicity and surface charge suggest that these chemical properties may impact the strength of the binding interactions and contribute to the differences in clinical efficacy among the IL-23 inhibitors. Further investigation to explore this potential correlation is needed.
In order to determine if each drug’s epitope SA correlated to the previously reported binding affinity and kinetic values, researchers employed linear regression analysis. They found that an inverse relationship existed between the epitope SA and the drugs’ dissociation equilibrium constant, suggesting that drugs with larger epitope SAs tend to exhibit higher binding affinities for IL-23. An inverse relationship was also noted between the epitope SA and the drugs’ dissociation rate constant, implying that drugs with larger epitope SAs may form more stable, longer-lasting, immune complexes.
Correlation of Epitope Properties and Short- and Long-Term Clinical Response
Based on their findings, the researchers suggested that drugs with more stable immune complexes with IL-23 would be more clinically efficacious for the treatment of plaque psoriasis. In order to test this theory, they utilized a meta-analysis to evaluate the comparative efficacy of IL-23 inhibitors in psoriasis. The drugs with highest efficacy rates (as measured by PASI-90 scores) in descending order were risankizumab, guselkumab, ustekinumab, tildrakizumab, with PASI (Psoriasis Area and Severity Index) defined as 90% or greater improvement from baseline.
When comparing this data with the structural properties for the epitopes, they found a positive correlation between epitope SA and short term (10-16 weeks) PASI-90 rates. In other words, drugs with larger epitope SAs demonstrated higher PASI-90 scores in the short term, suggesting better treatment outcomes.
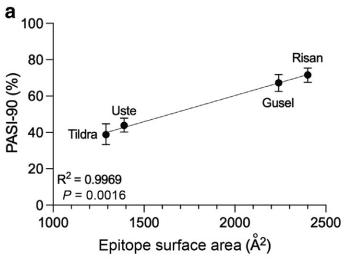
Increased binding affinity also positively correlated with higher PASI-90 scores as did the drugs’ dissociation rate constant. The drug’s association rate constant exhibited a negative correlation, however. This indicated that for anti-IL-23 biologics, that a slower dissociation of the biologic-IL-23 complex, rather than faster “on rate” or association of the complex, was the more important binding affinity component leading to higher treatment efficacy of psoriasis.
This positive correlation between epitope SA, binding affinity, dissociation kinetics, and clinical efficacy supports the initial theory of the researchers in suggesting that drugs which form more stable immune complexes with IL-23 are more likely to be effective in managing plaque psoriasis; in this study, risankizumab led the group.
To further evaluate the potential correlations between the structural properties of IL-23 inhibitors and their clinical efficacy, researchers utilized long-term clinical efficacy data from the same meta-analysis, excluding tildrakizumab as there was no long-term data available in that network meta-analysis at the time.
They found the same positive correlation between epitope SA, binding affinity, and dissociation rate constant, and long-term efficacy. There was also a negative trend with association constant and long-term efficacy, but the data was not statistically significant.
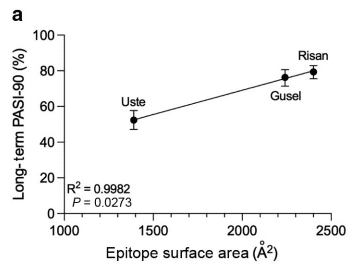
Researchers also noted that other epitope properties, such as total residue hydrophobicity, polarity, and charge content did not correlate with short- or long-term efficacy, implying that these particular chemical characteristics may not markedly impact the clinical performance of IL-23 inhibitors.
Exploring the Significance
The findings of this study reveal a significant positive correlation between the epitope SA of IL-23 inhibitors, their binding affinity, dissociation kinetics, and their clinical efficacy as measured by PASI-90 scores. This lends support to the hypothesis that the molecular and biochemical properties of these inhibitors may therefore directly influence patient outcomes and treatment responses. For dermatologists, this work importantly connects the molecular function of therapeutics with clinically relevant patient outcomes.
Of all the drug properties examined in this study, epitope SA appears to be the most significant in terms of correlation with clinical efficacy of the drugs. The association dynamics between the inhibitors and IL-23, by contrast, seem to be less important as it relates to clinical efficacy.
The positive association between epitope SA and efficacy seems to suggest that epitope SA majorly impacts stable drug-target complex formation and prevents biologic-IL-23 dissociation. Furthermore, the study reveals that direct epitope overlap with the IL-23R binding site is not required for clinical efficacy, as demonstrated by risankizumab and tildrakizumab. For some anti-IL-23 biologics, efficacy is linked to steric hindrance, which limits access of IL-23R to IL-23.
This study has significantly deepened our molecular understanding of IL-23 inhibitors and provides valuable information for researchers and clinicians. It provides a molecular rationale for drug selection and optimization strategies. Moreover, it demonstrates that despite targeting the same cytokine, IL-23 inhibitors do have distinct structural properties and molecular mechanisms of action, and therefore the clinical response to one drug may not necessarily indicate a clinical response to others within the same drug class.
While this study’s results have significantly advanced our knowledge of the molecular structures of IL-23 inhibitors, the authors note that our understanding can be expanded on in the future with additional detail. They add that information collected via x-ray crystallography or cryo-electron microscopy, for example, would better inform future studies and further improve our molecular understanding of these drugs.
Source:
- Structural Basis for p19 Targeting by Anti-IL-23 Biologics: Correlations with Short- and Long-Term Efficacy in Psoriasis
- Focus: Skin: Structural Basis for How Biologic Medicines Bind their Targets in Psoriasis Therapy – PMC (nih.gov)
- Armstrong AW, Puig L, Joshi A, et al. Comparison of Biologics and Oral Treatments for Plaque Psoriasis: A Meta-analysis . JAMA Dermatol. 2020;156(3):258–269. doi:10.1001/jamadermatol.2019.4029